How can the CCB enable your research?
The CCB assists researchers in any phase of their project, from basic questions about their biological systems, identifying therapeutic compounds, or improving existing hit compounds. Below are some examples of how the CCB helped researchers.
Probing the basic biology of your system
Predicting protein function of an effector proteinSidI was an effector protein with unknown structure and function. We employed de novo modeling combined with a structure similarity search to predict it to be a glycosyltransferase. This prediction was validated experimentally. Read about it here. | 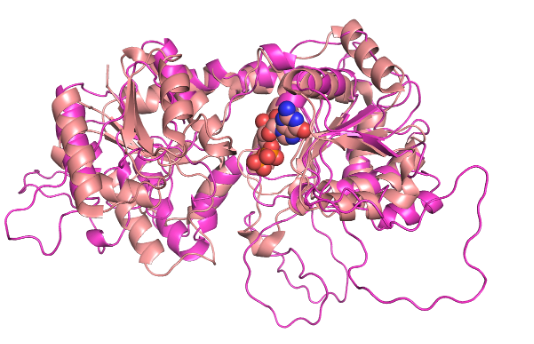 |
Modeling interactions of type III secretion system proteinsSctD and SctK interact to form part of the T3SS sorting platform. Protein-protein docking followed by computational alanine scanning was used to generate hypotheses of key interacting residues, which were experimentally validated. Read about it here. | 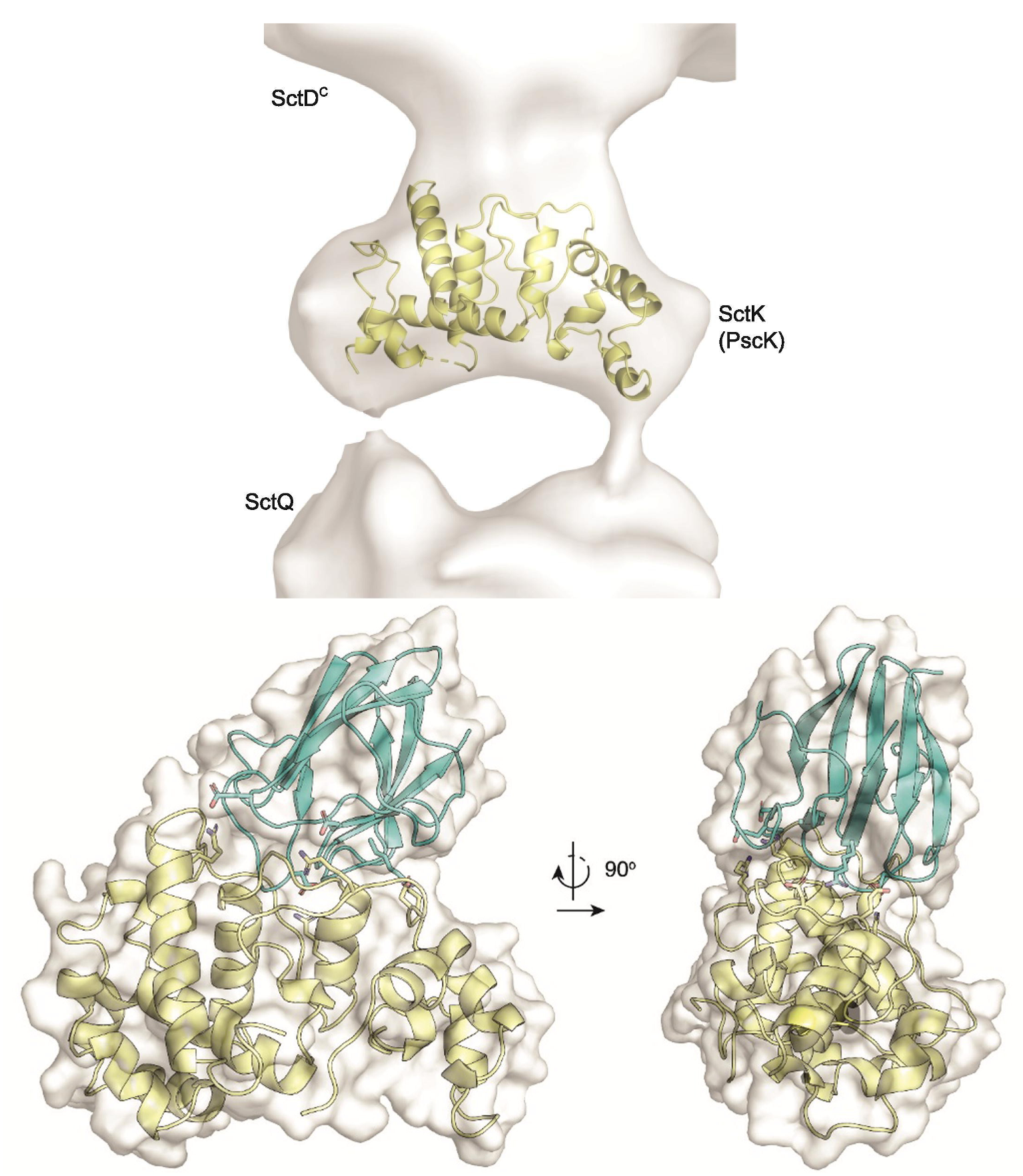 |
Assessing the effect of clinically relevant mutationsTRIM32 is an E3 ubiquitin ligase that contains a six-bladed beta-propeller domain, and two different point mutations are associated with limb-girdle muscular dystrophy. Rosetta Relax was used to assess the effect of each point mutation. The severity of the phenotype was found to match the scope of the disruption of the beta-propeller. Read about it here. | 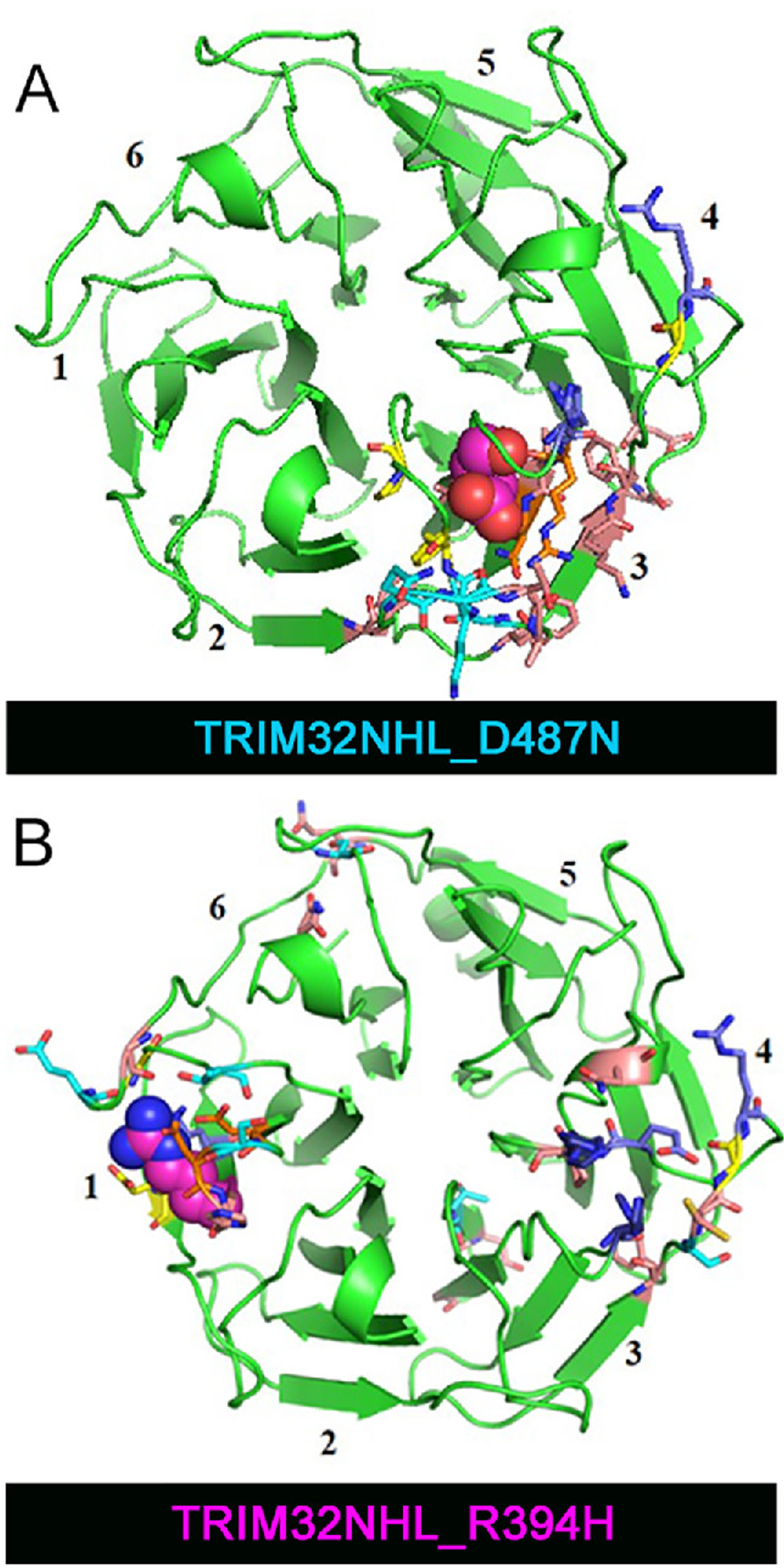 |
Assessing the binding and activity of peptidesBifunctional delta- and mu-opoid agonists such as endogenous enkephalins offer the potential to treat both acute and chronic pain, but also recruit beta-arresting, which cause adverse effects. We used Schrodinger Glide peptide docking to model a series of enkephalin derivatives to suggest key interactions involved in beta-arresting recruitment and explain differences in beta-arresting recruitment among the series. Read about it here. | 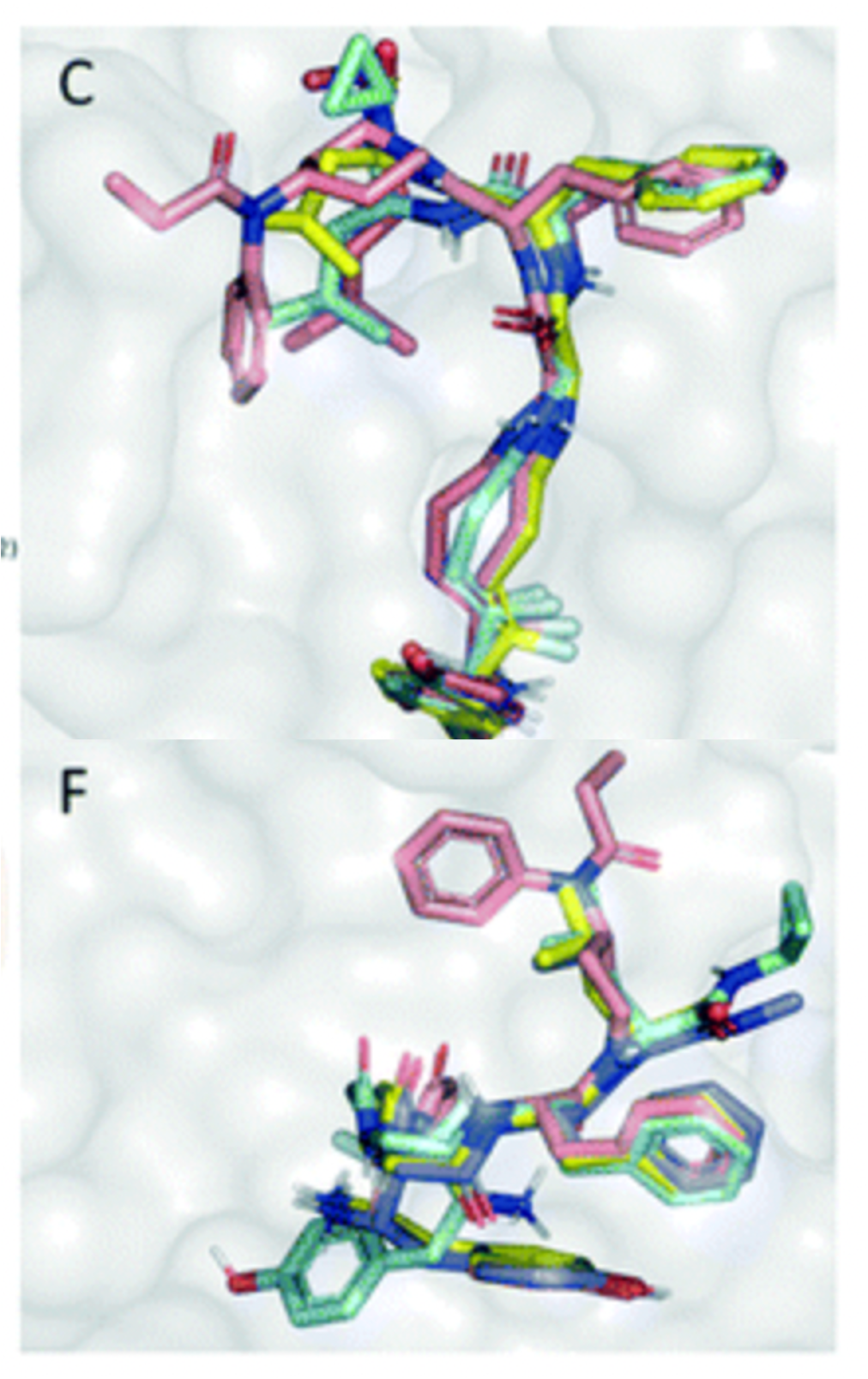 |
Identification of new therapeutic/probe compounds
Virtual screening for novel inhibitorsOur collaborators wanted a new ACMSD inhibitor. We performed a virtual screen using Schrodinger Glide. We identified diflunisal, an FDA approved drug, as an ACMDS inhibitor Its activity was validated through a structure-activity relationship campaign and X-ray crystallography. Read about it here. | 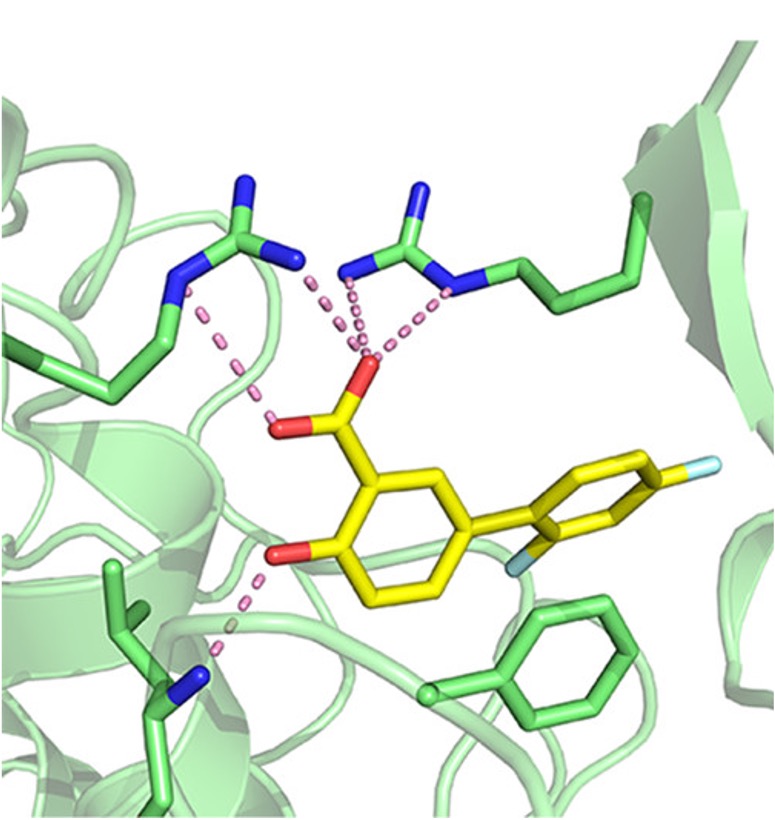 |
Hit-to-lead optimization
Cheminformatics analysis of high-throughput screening hitsEvery high-throughput screening needs a cheminformatics analysis. By performing clustering, medicinal chemistry filtering, and physiochemical property analysis, we can help weed out false positives and help select the best compounds for hit-to-lead optimization. See an example here. | 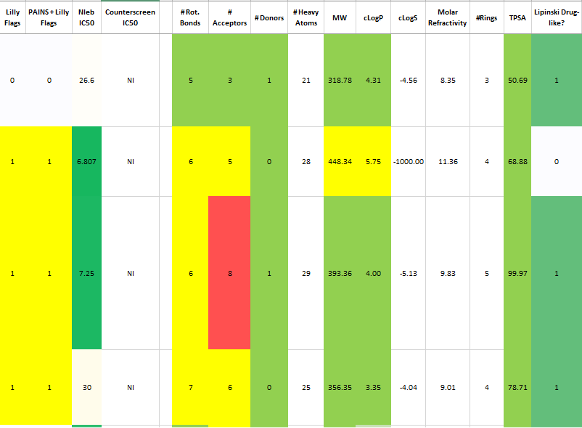 |
Structure guided optimization of a reversible inhibitorOur collaborators sought to improve the potency of a bicyclic hepatitis C NS5A genotype 1A inhibitor. We used template-based docking to model the dimer in a closed form, and used Glide to identify a pose that was consistent with site directed mutagenesis studies. Using this model, we proposed analogs that would take advantage of a nearby hydrophobic cleft. Read about it here. |  |
Optimization of a covalent inhibitorIlludalic acid is an inhibitor of the receptor tyrosine phosphatase PTPRD that covalently attaches to the catalytic cysteine. We developed a protocol to use Schrodinger Glide to quickly screen analogs, and then model the covalent bond to select compounds for synthesis. Using an iterative process of design, docking, synthesis, and testing, we identified more potent PTPRD inhibitors. Read about it here. | 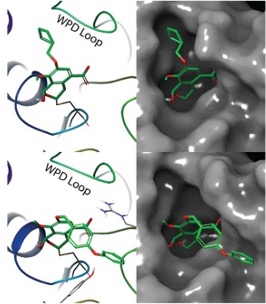 |